Automated Deep Learning for Biomedical Image Analysis
summary
AIxCell is an innovative research project designed to revolutionize the analysis of biomedical image data, such as cell microscopy, by automating traditionally manual and expertise-intensive processes. The project utilizes advanced deep learning (DL) techniques to develop an AutoML software tool, enabling biomedical professionals—including biologists, medical practitioners, and virologists—to independently create and apply deep learning solutions tailored to their specific challenges.
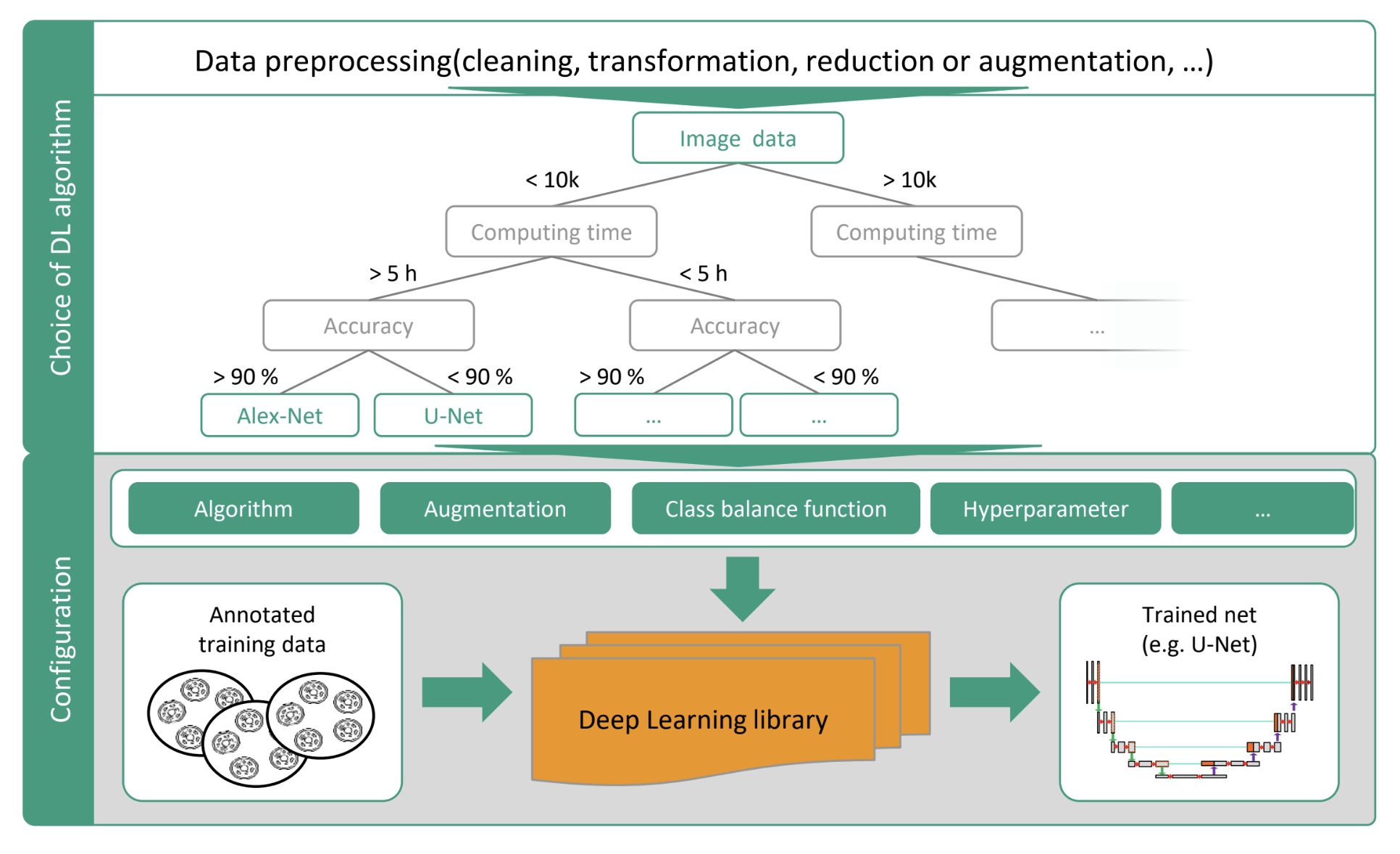
problem statement
Biomedical image analysis is currently labor-intensive, requiring extensive expertise and resources. The process is often subjective and prone to device- and application-specific errors, compromising result comparability. Outsourcing custom deep learning (DL) solutions is often cost-prohibitive for many institutions, and non-experts lack the technical skills to develop these tools independently. These challenges highlight the need for a solution that simplifies and democratizes access to DL technologies.
objectives
The primary goal of AIxCell was to enable domain experts to autonomously train and deploy deep learning (DL) models for their specific use cases, without requiring extensive expertise in machine learning or software development. This was achieved through:
- Development of a domain-specific meta-learning system.
- Creation of an AutoML software tool that selects optimal DL algorithms, covering preprocessing, model selection, configuration, and postprocessing.
- Integration of these tools into a user-friendly application with functionalities for dataset annotation, task specification, result visualization, and DL model utilization.
key innovations
- Meta-Learning System (AutoKonfig):
- A core component of AIxCell, this decision logic system selects the most suitable algorithm configurations based on input data, application requirements, and available resources.
- Utilizes a library of pre-trained deep learning (DL) algorithms, datasets, requirements, and evaluation results to make informed testimonials.
- The system continuously learns and refines its algorithm selection process for specific tasks.
- Deep Learning Algorithm Library:
- A comprehensive repository of developed algorithms, accompanied by metadata such as datasets used, evaluation metrics, and resource requirements.
- Serves as the foundation for training the Meta-Learning System.
- User-Friendly Application:
- Provides an intuitive interface that allows biomedical experts to input data, specify tasks, review results, and utilize trained models—no technical expertise required.
outcomes
AIxCell successfully demonstrated how automation can transform biomedical image analysis by reducing reliance on external expertise and lowering costs. It provided a scalable solution for creating high-quality deep learning (DL) applications tailored to specific biomedical challenges.
funding & collaboration
The project was funded through the AiF under the Industrial Collective Research (IGF) program by the German Federal Ministry for Economic Affairs and Climate Action, with the funding reference number 21361 N. AIxCell was developed in collaboration with various partners from research and industry.
master’s thesis contribution
As part of the AIxCell project, I authored my master’s thesis titled “Benchmarking of Deep Learning Algorithms for Stem Cell Classification” (download here), where I developed a modular deep learning pipeline for cell culture analysis, focusing on investigating the proliferation of cardiomyocytes.
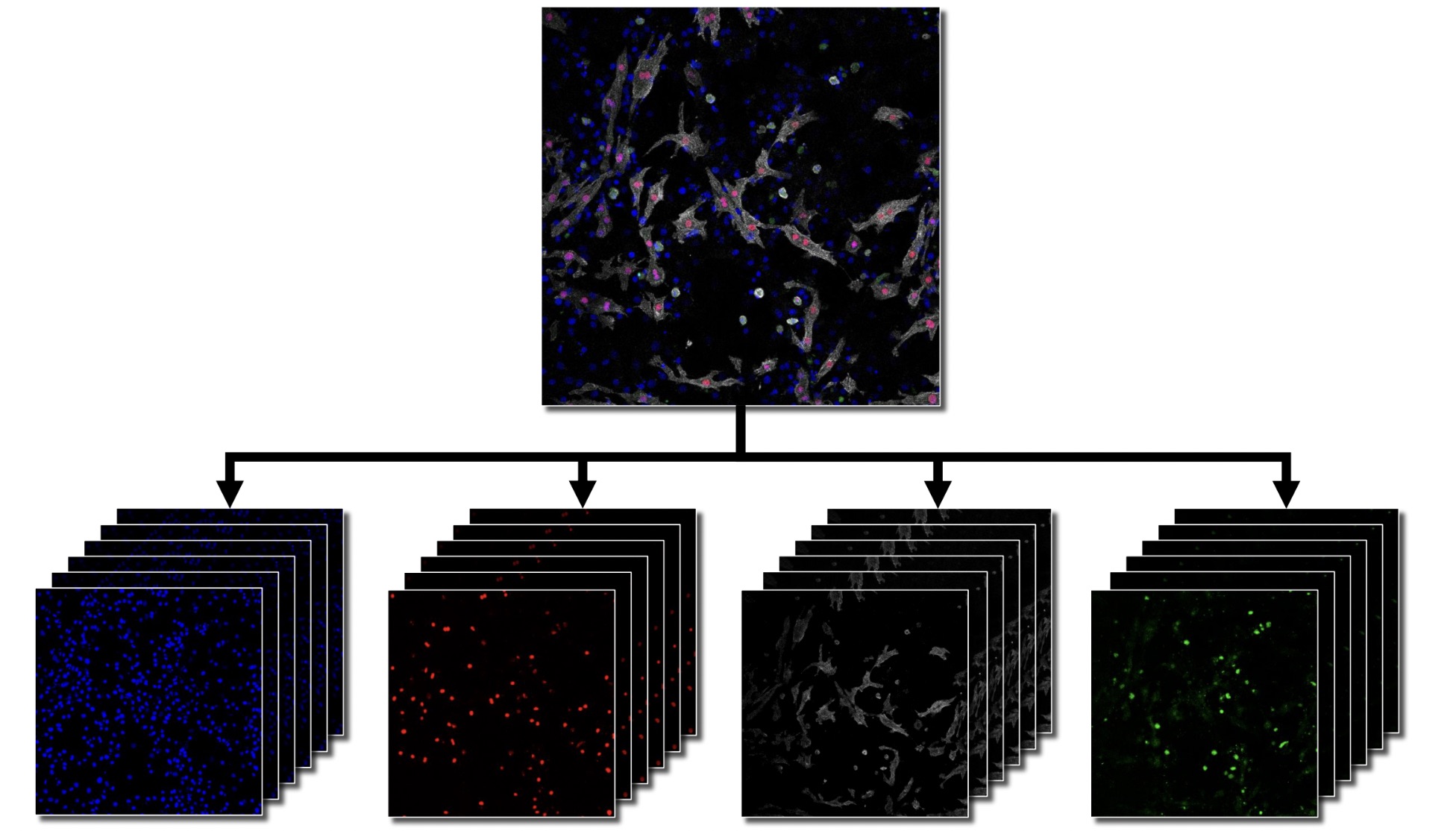
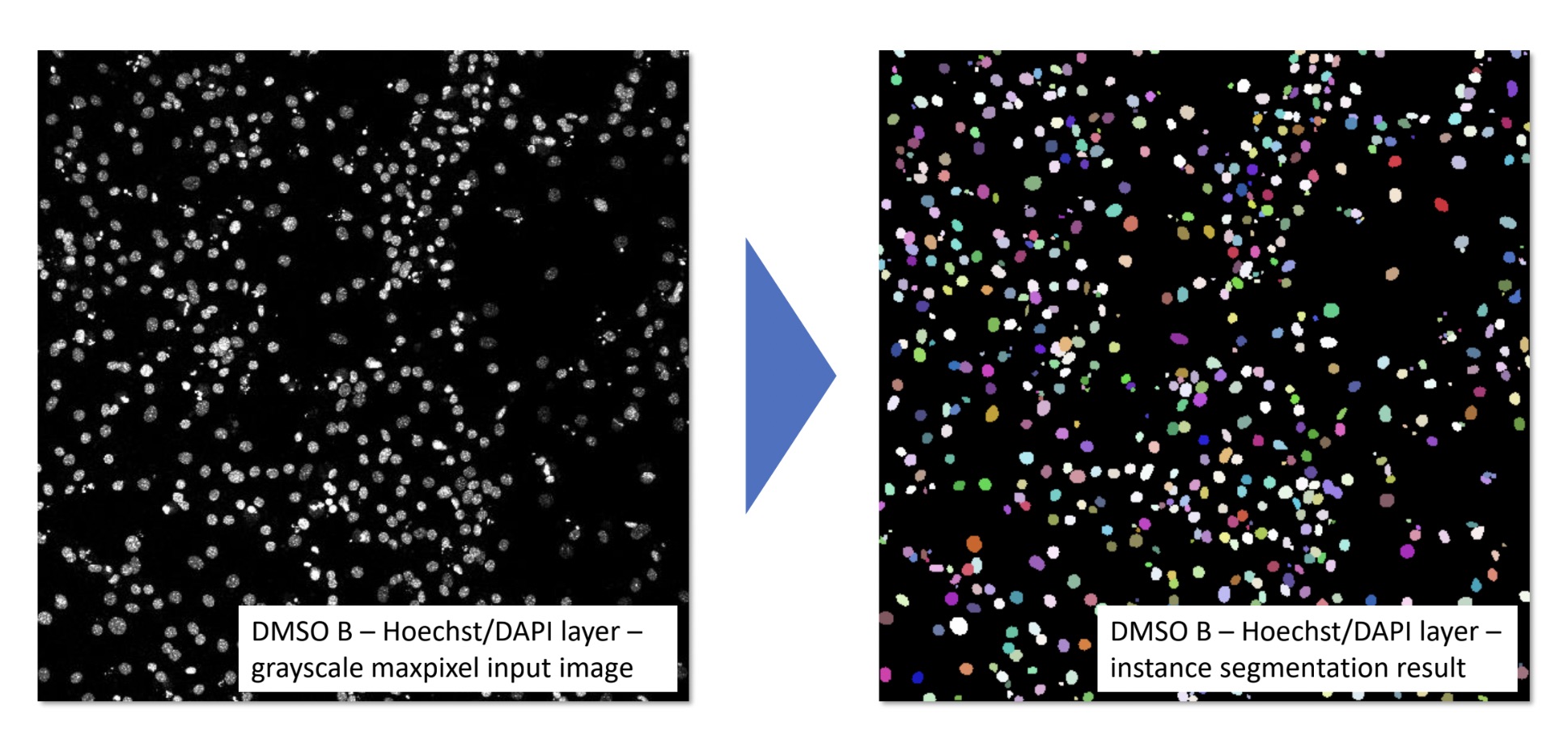
In this thesis, I designed and implemented a flexible, three-stage deep learning-based image analysis pipeline for the classification of mononuclear and binuclear cardiomyocytes in confocal microscopy data. The pipeline features a modular architecture with distinct preprocessing, modeling, and postprocessing stages, ensuring adaptability to various imaging modalities and cell analysis tasks. This approach provides robustness and scalability, making it suitable for broader applications in cell culture research.
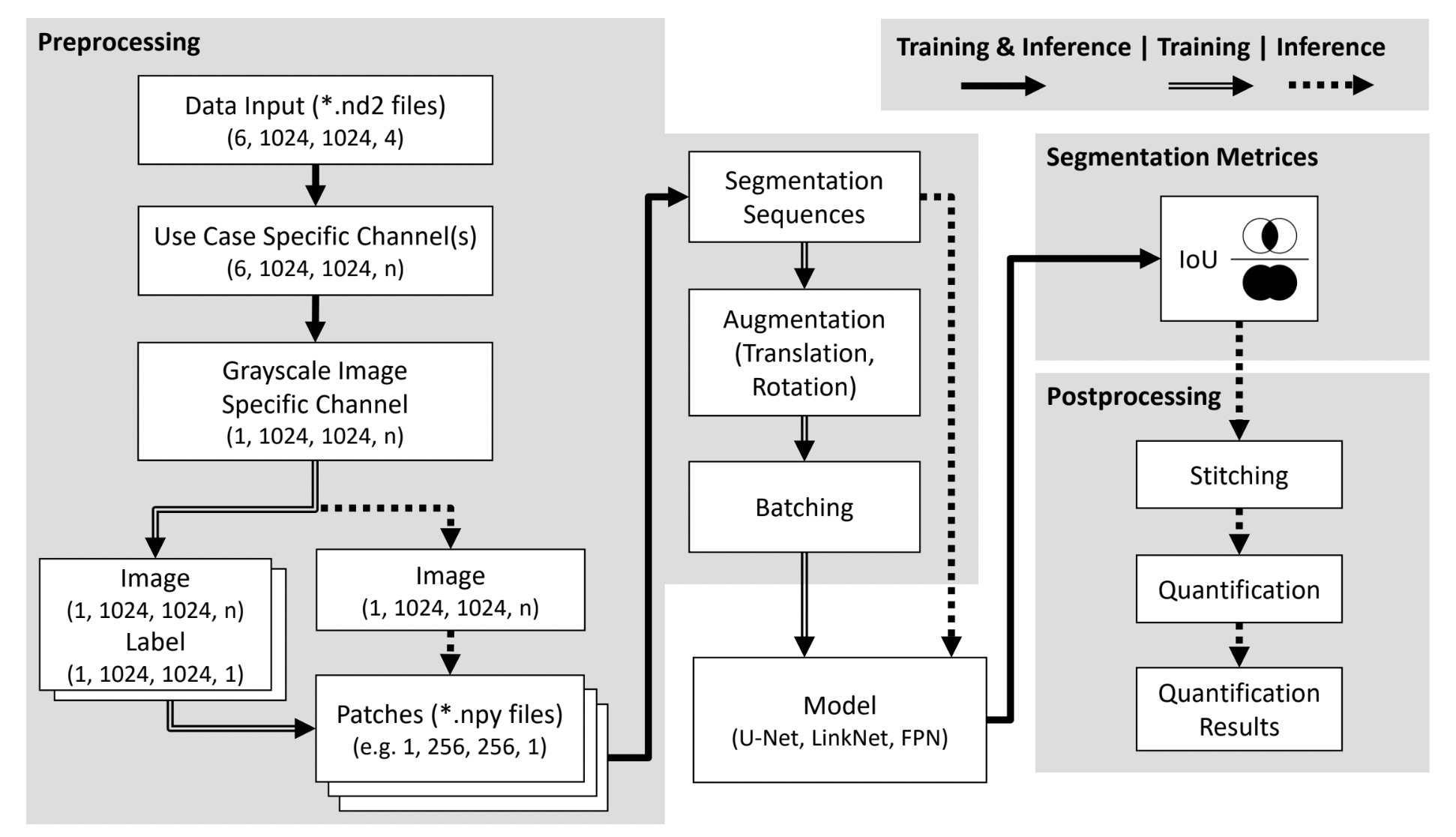
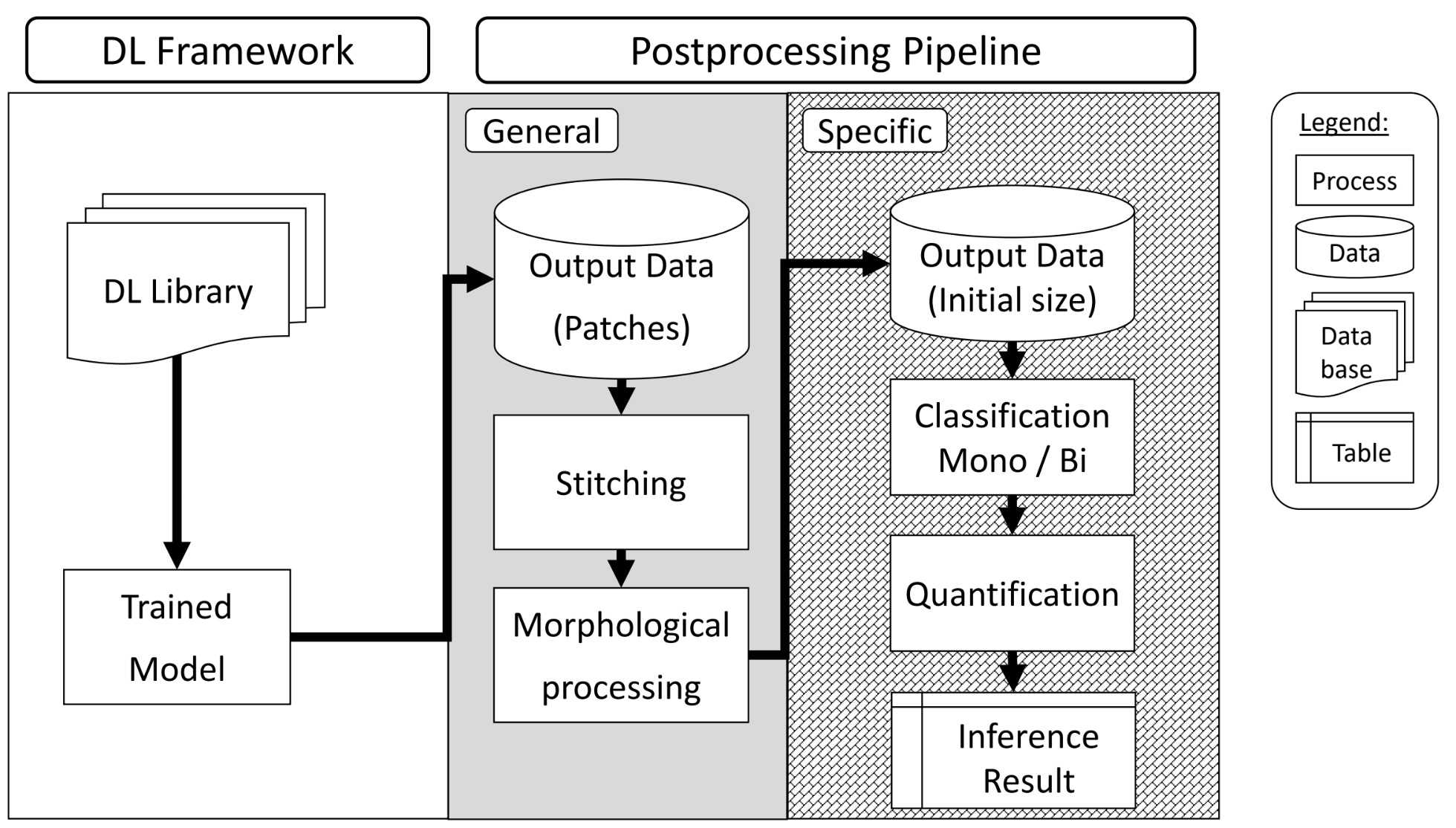
key aspects of the thesis include:
- Development of a data-efficient and flexible pipeline for analyzing cardiomyocytes (CMs) in complex multi-cell classification scenarios.
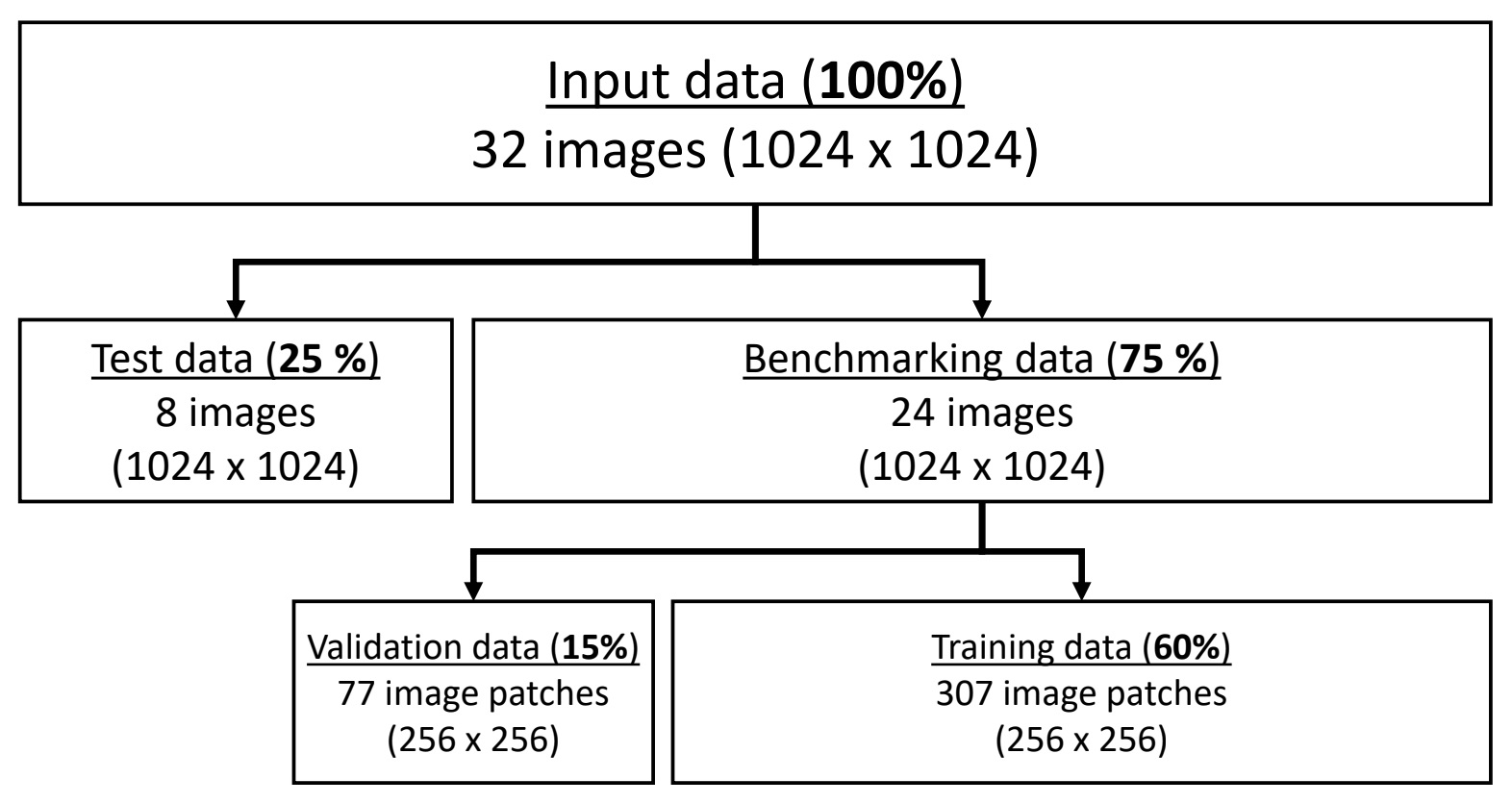
- Benchmarking of 18 different encoder-decoder model architectures for semantic segmentation.
- Conducting 173 experiments, including hyperparameter optimization and evaluation of dataset-related effects.
- Demonstrating the pipeline’s ability to perform well even with small datasets, a critical factor for laboratory-specific analyses.
- Achieving high test accuracies (up to 82%) for the classification of nucleus types in cardiomyocytes.
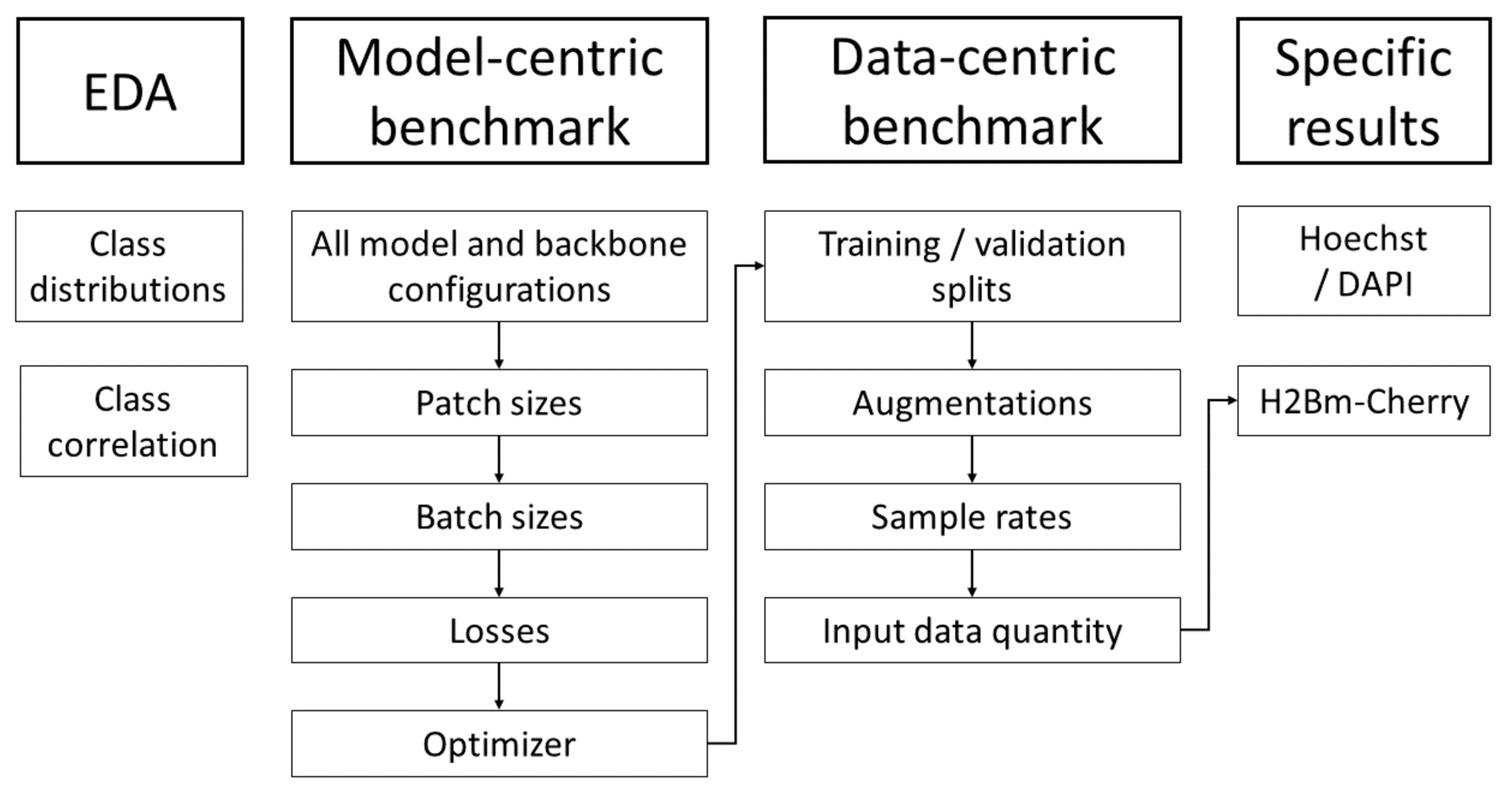
The findings from this thesis were further published as a research paper (download here) at the MIDL2022 conference in Zürich, Switzerland, contributing valuable insights into automated biomedical image analysis using deep learning techniques.
This thesis contributes to the broader goals of the AIxCell project by benchmarking different deep learning (DL) algorithms for a specific biomedical use case: segmenting and classifying mono- and binuclear cardiomyocytes in confocal microscopy images. This analysis is aimed at assessing the influence of various substances on the proliferation of heart tissue in mice. The thesis evaluates how different DL models perform in this context, providing insights into the most effective approaches for automated image analysis in cardiomyocyte research.
For more information about AIxCell check the links below.
relevant links
- Fraunhofer IPT - project page
- Fraunhofer IPT - press release
- F.O.M. - project page
- F.O.M. - press release
- KI-LAB Bodensee
- Lernende Systeme
- Masters thesis
- Peer-reviewed publication
I participated in this project as part of my master's thesis at the Fraunhofer Institute for Production Technology in Aachen, Germany.